En palabras simples, la prueba de hipótesis de permutación en R es una forma de comparar un valor numérico de 2 grupos. La prueba de hipótesis de permutación es una alternativa a:
- Prueba t independiente de dos muestras
- Prueba de suma de rangos de Mann-Whitney U, también conocida como Wilcoxon
Implementemos esta prueba en la programación R.
¿ Por qué utilizar la Prueba de Hipótesis de Permutación?
- Tamaño de muestra pequeño.
- No se cumplen los supuestos (para el enfoque paramétrico).
- Pruebe algo diferente a los enfoques clásicos comparando Medias y Medianas.
- Difícil estimar el SE para la estadística de prueba.
Pasos de la prueba de hipótesis de permutación
- Especificar una hipótesis
- Elija test-stat (por ejemplo: media, mediana, etc.)
- Determinar la distribución de test-stat
- Convertir test-stat a valor P
Nota:
Valor P = N.º de permutaciones que tienen un valor de prueba-estado mayor que el valor de prueba-estado observado/N.º de permutaciones.
Implementación en R
- Conjunto de datos: la descarga
- Hipótesis: El peso del
Test-Estadísticas
- diferenciaindependientedos muestras
- Test-Statistics #2: El valor absoluto de la diferencia en los pesos medianos para las dos dietas
R
# R program to illustrate
# Permutation Hypothesis Test
# load the data set
d <- read.table(file = "ChickData.csv",
header = T, sep = ",")
# print the dataset
print(d)
# check the names
names(d)
levels(d$feed)
# how many observations in each diet?
table(d$feed)
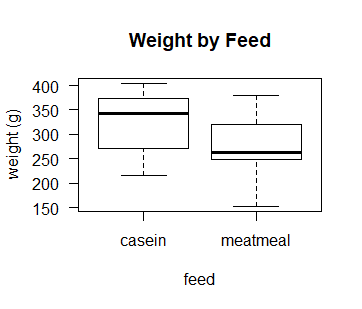
# let's look at a boxplot of weight gain by those 2 diets
boxplot(d$weight~d$feed, las = 1,
ylab = "weight (g)",
xlab = "feed",
main = "Weight by Feed")
# calculate the difference in sample MEANS
mean(d$weight[d$feed == "casein"]) # mean for casein
mean(d$weight[d$feed == "meatmeal"]) # mean for meatmeal
# lets calculate the absolute diff in means
test.stat1 <- abs(mean(d$weight[d$feed == "casein"]) -
mean(d$weight[d$feed == "meatmeal"]))
test.stat1
# calculate the difference in sample MEDIANS
median(d$weight[d$feed == "casein"]) # median for casein
median(d$weight[d$feed == "meatmeal"]) # median for meatmeal
# lets calculate the absolute diff in medians
test.stat2 <- abs(median(d$weight[d$feed == "casein"]) -
median(d$weight[d$feed == "meatmeal"]))
test.stat2
# Permutation Test
# for reproducability of results
set.seed(1979)
# the number of observations to sample
n <- length(d$feed)
# the number of permutation samples to take
P <- 100000
# the variable we will resample from
variable <- d$weight
# initialize a matrix to store the permutation data
PermSamples <- matrix(0, nrow = n, ncol = P)
# each column is a permutation sample of data
# now, get those permutation samples, using a loop
# let's take a moment to discuss what that code is doing
for(i in 1:P)
{
PermSamples[, i] <- sample(variable,
size = n,
replace = FALSE)
}
# we can take a quick look at the first 5 columns of PermSamples
PermSamples[, 1:5]
# initialize vectors to store all of the Test-stats
Perm.test.stat1 <- Perm.test.stat2 <- rep(0, P)
# loop thru, and calculate the test-stats
for (i in 1:P)
{
# calculate the perm-test-stat1 and save it
Perm.test.stat1[i] <- abs(mean(PermSamples[d$feed == "casein",i]) -
mean(PermSamples[d$feed == "meatmeal",i]))
# calculate the perm-test-stat2 and save it
Perm.test.stat2[i] <- abs(median(PermSamples[d$feed == "casein",i]) -
median(PermSamples[d$feed == "meatmeal",i]))
}
# before going too far with this,
# let's remind ourselves of
# the TEST STATS
test.stat1; test.stat2
# and, take a look at the first 15
# permutation-TEST STATS for 1 and 2
round(Perm.test.stat1[1:15], 1)
round(Perm.test.stat2[1:15], 1)
# and, let's calculate the permutation p-value
# notice how we can ask R a true/false question
(Perm.test.stat1 >= test.stat1)[1:15]
# and if we ask for the mean of all of those,
# it treats 0 = FALSE, 1 = TRUE
mean((Perm.test.stat1 >= test.stat1)[1:15])
# Calculate the p-value, for all P = 100,000
mean(Perm.test.stat1 >= test.stat1)
# and, let's calculate the p-value for
# option 2 of the test statistic (abs diff in medians)
mean(Perm.test.stat2 >= test.stat2)
Producción:
> print(d)
weight feed
1 325 meatmeal
2 257 meatmeal
3 303 meatmeal
4 315 meatmeal
5 380 meatmeal
6 153 meatmeal
7 263 meatmeal
8 242 meatmeal
9 206 meatmeal
10 344 meatmeal
11 258 meatmeal
12 368 casein
13 390 casein
14 379 casein
15 260 casein
16 404 casein
17 318 casein
18 352 casein
19 359 casein
20 216 casein
21 222 casein
22 283 casein
23 332 casein
> names(d)
[1] "weight" "feed"
> levels(d$feed)
[1] "casein" "meatmeal"
> table(d$feed)
casein meatmeal
12 11
> mean(d$weight[d$feed == "casein"]) # mean for casein
[1] 323.5833
> mean(d$weight[d$feed == "meatmeal"]) # mean for meatmeal
[1] 276.9091
> test.stat1
[1] 46.67424
> median(d$weight[d$feed == "casein"]) # median for casein
[1] 342
> median(d$weight[d$feed == "meatmeal"]) # median for meatmeal
[1] 263
> test.stat2
[1] 79
> PermSamples[, 1:5]
[,1] [,2] [,3] [,4] [,5]
[1,] 379 283 380 352 206
[2,] 380 303 258 260 380
[3,] 257 206 379 380 153
[4,] 283 242 222 404 359
[5,] 222 260 325 258 258
[6,] 315 352 153 379 263
[7,] 352 263 263 325 325
[8,] 153 325 315 359 216
[9,] 368 379 344 242 260
[10,] 344 258 368 368 257
[11,] 359 257 206 257 315
[12,] 206 153 404 222 303
[13,] 404 344 303 390 390
[14,] 325 318 318 303 352
[15,] 242 404 332 263 404
[16,] 390 380 257 206 379
[17,] 260 332 216 315 318
[18,] 303 359 352 344 368
[19,] 263 222 242 283 222
[20,] 332 368 260 332 344
[21,] 318 315 283 318 283
[22,] 216 390 390 153 332
[23,] 258 216 359 216 242
> test.stat1; test.stat2
[1] 46.67424
[1] 79
> round(Perm.test.stat1[1:15], 1)
[1] 17.1 32.4 17.6 47.1 56.1 28.9 31.0 40.8 6.8 13.8 9.1 46.5 28.9 50.9 32.7
> round(Perm.test.stat2[1:15], 1)
[1] 61.0 75.0 4.5 59.0 78.0 17.0 62.0 38.5 4.5 16.0 23.0 60.5 63.5 75.0 37.0
> (Perm.test.stat1 >= test.stat1)[1:15]
[1] FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
> mean((Perm.test.stat1 >= test.stat1)[1:15])
[1] 0.2
> mean(Perm.test.stat1 >= test.stat1)
[1] 0.09959
> mean(Perm.test.stat2 >= test.stat2)
[1] 0.05407
Publicación traducida automáticamente
Artículo escrito por samrat2825 y traducido por Barcelona Geeks. The original can be accessed here. Licence: CCBY-SA